PvuI-HF®
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
R3150S |
PvuI-HF |
500 units ( 20000 units/ml ) | - | Unavailable in your region | |
R3150L |
PvuI-HF |
2.500 units ( 20000 units/ml ) | - | Unavailable in your region |
PvuI-HF®
PvuI-HF has been reformulated with Recombinant Albumin (rAlbumin) beginning with Lot #10128368. Learn more

Product Introduction
High-Fidelity (HF®) restriction enzymes have the same specificity as native enzymes, but have been engineered for significantly reduced star activity and performance in a single buffer (rCutSmart™ Buffer). All HF-restriction enzymes come with Gel Loading Dye, Purple (6X). Enjoy the enhanced performance and added value of our engineered enzymes at the same price as the native enzyme:
- Engineered for improved performance
- 100% activity in rCutSmart Buffer
- Time-Saver™ qualified for digestion in 5-15 minutes
- Reduced star activity
- Supplied with 1 vial of Gel Loading Dye, Purple (6X)
- Restriction Enzyme Cut Site: CGAT/CG
| Catalog # | Size | Concentration |
|---|---|---|
| R3150S | 500 units | 20000 units/ml |
| R3150L | 2500 units | 20000 units/ml |
Featured Videos
View Video Library-

Reduce Star Activity with High-Fidelity Restriction Enzymes
-

TIME-SAVER™ Protocol for Restriction Enzyme Digests
-

NEB® TV Ep. 15 – Applications of Restriction Enzymes
-

Restriction Enzyme Digest Protocol: Cutting Close to DNA End
-

Restriction Enzyme Digestion Problem: DNA Smear on Agarose Gel
-

Why is My Restriction Enzyme Not Cutting DNA?
-

Restriction Enzyme Digest Problem: Too Many DNA Bands
-

Double Digestion with NEBcloner
- Product Information
- Protocols, Manuals & Usage
- Tools & Resources
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
High Fidelity (HF) Restriction Enzymes have 100% activity in rCutSmart Buffer; single-buffer simplicity means more straightforward and streamlined sample processing. HF enzymes also exhibit dramatically reduced star activity. HF enzymes are all Time-Saver qualified and can therefore cut substrate DNA in 5-15 with the flexibility to digest overnight without degradation to DNA. Engineered with performance in mind, HF restriction enzymes are fully active under a broader range of conditions, minimizing off-target products, while offering flexibility in experimental design.NEB extensively performs quality controls on all standard and high-fidelity (HF) restriction enzymes. Examples of nuclease contamination studies for some of our HF restriction enzymes are shown below.
Restriction Enzyme Competitor Study: Nuclease Contamination
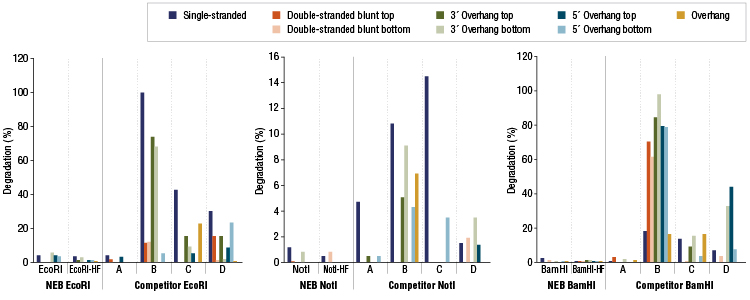
EcoRI, NotI, and BamHI from multiple suppliers were tested in reactions containing a fluorescent labeled single stranded, double stranded blunt, 3’overhang or 5’ overhang containing oligonucleotides. The percent degradation is determined by capillary electrophoresis and peak analysis. The resolution is at the single nucleotide level.
Product Source
An E. coli strain that carries the cloned and modified PvuI gene from Proteus vulgaris (ATCC 13315).- This product is related to the following categories:
- Restriction Endonucleases P R,
- High-Fidelity (HF®) Restriction Endonucleases,
- Time-Saver Qualified Restriction Enzymes,
- This product can be used in the following applications:
- Fast Cloning: Accelerate your cloning workflows with reagents from NEB,
- Restriction Enzyme Digestion
Reagents Supplied
Reagents Supplied
The following reagents are supplied with this product:
| NEB # | Component Name | Component # | Stored at (°C) | Amount | Concentration | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
Properties & Usage
Unit Definition
One unit is defined as the amount of enzyme required to digest 1 µg of pXba DNA in 1 hour at 37°C in a total reaction volume of 50 µl.Reaction Conditions
1X rCutSmart™ Buffer
Incubate at 37°C
1X rCutSmart™ Buffer
50 mM Potassium Acetate
20 mM Tris-acetate
10 mM Magnesium Acetate
100 µg/ml Recombinant Albumin
(pH 7.9 @ 25°C)
Activity in NEBuffers
NEBuffer™ r1.1: 25%NEBuffer™ r2.1: 100%
NEBuffer™ r3.1: 100%
rCutSmart™ Buffer: 100%
Diluent Compatibility
Storage Buffer
10 mM Tris-HCl
300 mM NaCl
1 mM DTT
0.1 mM EDTA
500 µg/ml Recombinant Albumin
50% Glycerol
pH 7.4 @ 25°C
Heat Inactivation
NoMethylation Sensitivity
dam methylation: Not Sensitive
dcm methylation: Not Sensitive
CpG Methylation: Blocked
Related Products
Materials Sold Separately
Product Notes
- Cleavage of mammalian genomic DNA is blocked by CpG methylation.
- For enzymes that cannot be heat-inactivated, we recommend using a column for cleanup (such as the Monarch® PCR & DNA Cleanup Kit), or running the reaction on an agarose gel and then extracting the DNA (we recommend Monarch Gel Extraction Kit), or performing a phenol/chloroform extraction.
Protocols, Manuals & Usage
Protocols
Usage & Guidelines
- Activity at 37°C for Restriction Enzymes with Alternate Incubation Temperatures
- Activity of Restriction Enzymes in PCR Buffers
- Cleavage Close to the End of DNA Fragments
- Dam and Dcm Methylases of E. coli
- Digestion of Agarose-Embedded DNA: Info for Specific Enzymes
- Double Digests
- Effects of CpG Methylation on Restriction Enzyme Cleavage
- Heat Inactivation
- NEBuffer Activity/Performance Chart with Restriction Enzymes
- Optimizing Restriction Endonuclease Reactions
- Restriction Endonucleases - Survival in a Reaction
- Restriction Enzyme Diluent Buffer Compatibility
- Restriction Enzyme Tips
- Single Letter Codes
- Star Activity
- Traditional Cloning Quick Guide
Tools & Resources
Selection Charts
- Alphabetized List of Recognition Sequences
- Cleavage of Supercoiled DNA
- Compatible Cohesive Ends and Generation of New Restriction Sites
- Dam-Dcm and CpG Methylation
- Frequencies of Restriction Sites
- Isoelectric Points (pI) for Restriction Enzymes
- Isoschizomers
- NEB Diluent and Buffer Table
- Time-Saver™ Qualified Enzymes
- Why Choose Recombinant Enzymes?
Web Tools
FAQs & Troubleshooting
FAQs
- How can I search for a restriction enzyme by sequence, overhang or name?
- How should I stop my restriction digest?
- How stable is a particular restriction enzyme?
- When should I choose the HF version of the enzyme?
- When is star activity a concern?
- How should I set up a restriction digest?
- I don't see any cleavage after my restriction digest. What factors can interfere with cleavage?
- How can I generate a restriction enzyme site map for my sequence?
- What information is available in the Restriction Enzyme Database (REBASE)?
- Is extended digestion (incubation times > 1 hour) recommended?
- Do degenerate recognition sites need to be palindromic?
- What does HF® refer to following the name of a restriction enzyme?
- Do I have to set-up digests with Time-Saver™ qualified enzymes for 5-15 minutes? Can I digest longer?
- Can Gel Loading Dye, Purple 6X (B7024) be stored in cold temperatures?
- What does it mean to be Time-Saver™ qualified?
- Which NEB restriction enzymes are supplied with Gel Loading Dye, Purple (6X)?
- Is this enzyme sensitive to dam, dcm or mammalian CpG methylation?
- Can you tell me more about the switch from BSA to Recombinant Albumin (rAlbumin) in NEBuffers?
Troubleshooting
Citations & Technical Literature
Citations
Additional Citations
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specification Change Notifications
Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- R3150S_L_v1_0031312
- R3150S_L_v1_0021301
- R3150S_L_v1_0021306
- R3150S_L_v1_0031405
- R3150S_L_v1_0031412
- R3150S_L_v1_0031505
- R3150S_L_v1_0031510
- R3150S_L_v1_0031604
- R3150S_L_v1_0031610
- R3150S_L_v1_0031703
- R3150S_L_v1_0031708
- R3150S_L_v1_0031802
- R3150L_v1_10017176
- R3150S_v1_10017174
- R3150L_v1_10026318
- R3150S_v1_10026313
- R3150S_v1_10041275
- R3150L_v1_10044772
- R3150S_v1_10050239
- R3150S_v1_10055123
- R3150L_v1_10056492
- R3150L_v1_10061063
- R3150S_v1_10059902
- R3150L_v1_10067958
- R3150S_v1_10068967
- R3150L_v1_10071128
- R3150L_v1_10078386
- R3150S_v1_10079775
- R3150L_v1_10082756
- R3150S_v1_10083542
- R3150L_v1_10089671
- R3150S_v1_10089118
- R3150L_v1_10089115
- R3150S_v1_10089672
- R3150S_v1_10095186
- R3150L_v1_10098309
- R3150S_v1_10100681
- R3150L_v1_10107513
- R3150S_v1_10108027
- R3150L_v1_10119865
- R3150S_v1_10119205
- R3150L_v2_10128372
- R3150S_v2_10128371
- R3150S_v2_10138087
- R3150L_v2_10138086
- R3150S_v2_10151165
- R3150L_v2_10153273
- R3150S_v2_10160233
- R3150L_v2_10160235
- R3150L_v2_10171787
- R3150S_v2_10171786
- R3150S_v2_10179436
- R3150S_v2_10185769
- R3150L_v2_10179437
- R3150L_v2_10191121
- R3150S_v2_10191117
- R3150S_v2_10204455
- R3150L_v2_10200641
- R3150L_v2_10209956
- R3150S_v2_10218631
- R3150L_v2_10227934
- R3150L_v2_10235588
- R3150S_v2_10235510
- R3150S_v2_10244093
- R3150L_v2_10252816
- R3150S_v2_10264801
- R3150L_v2_10265622
- R3150L_v2_10279758
- R3150S_v2_10278732
- R3150S_v2_10287875
- R3150S_v2_10305912
- R3150L_v2_10305914
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.PvuI-HF®
rCutSmart™ Buffer
Gel Loading Dye, Purple (6X)
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email [email protected].This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.


















