PURExpress® In Vitro Protein Synthesis Kit
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
E6800S |
PURExpress In vitro Protein Synthesis Kit |
10 rxns | - | Unavailable in your region | |
E6800L |
PURExpress In vitro Protein Synthesis Kit |
100 rxns | - | Unavailable in your region |
PURExpress® In Vitro Protein Synthesis Kit
Product Introduction
PURExpress® is a reconstituted protein synthesis system based on the PUREsystem™ (Shimizu et al., 2001) where all necessary components needed for in vitro transcription and translation are purified from E. coli.• Defined system with all his-tagged proteins for coupled transcription/translation; Ribosome is not his-tagged
• T7 RNA Polymerase drives in vitro transcription
• Minimal nuclease and protease activity for stability of synthesized protein and encoding target
• Templates can be either plasmid DNA, linear DNA or mRNA
• Protein of interest can be synthesized and visualized in a few hours
• Synthesizes various target peptides and proteins
• Synthesized protein can be co-translationally radiolabeled or fluorescently labeled
• Protein can be reverse-purified or subject to direct functional analysis
• Applications include high throughput screening/directed evolution, synthetic biology, toxic or difficult to express protein synthesis, studies on protein folding, activity and protein-protein interactions
• Due to reconstituted nature, several kits are offered where translation factors or macromolecules have been omitted to facilitate specific studies (see companion products)
• Compatible with the PURExpress Disulfide Bond Enhancer (NEB #E6820)
| Catalog # | Size | Concentration |
|---|---|---|
| E6800S | 10.0 reactions | |
| E6800L | 100.0 reactions |
- Product Information
- Protocols, Manuals & Usage
- Tools & Resources
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
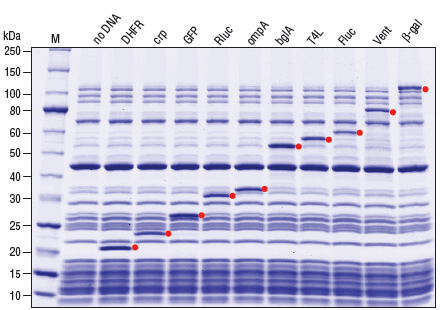
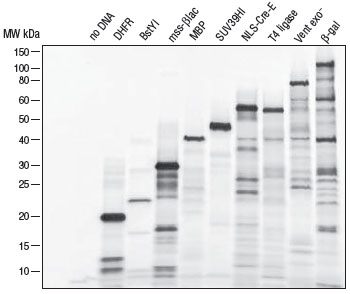
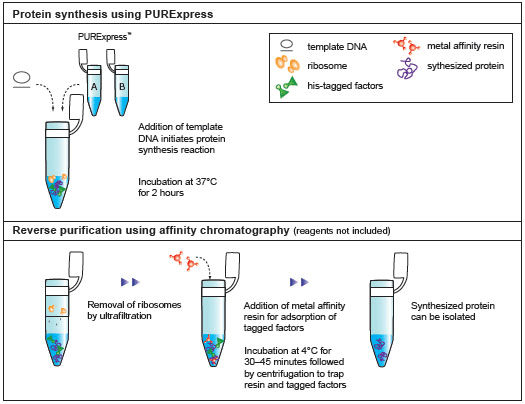
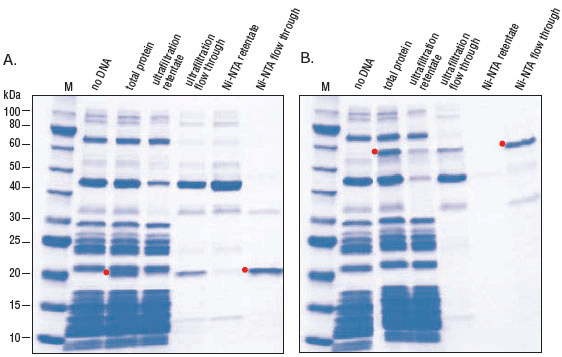
A rapid method for gene expression analysis, PURExpress® is a novel cell-free transcription/translation system reconstituted from the purified components necessary for E. coli translation. With minimal nuclease and protease activity, the PURExpress system preserves the integrity of DNA and RNA templates/complexes and results in proteins that are free of modification and degradation. Transcription and translation are carried out in a one-step reaction, and require the mixing of only two tubes. With results available in a few hours, PURExpress saves valuable laboratory time and is ideal for high throughput technologies.PURExpress Citations
- This product is related to the following categories:
- PURExpress,
- Cell-Free Protein Expression Products,
- Protein Expression Products,
- This product can be used in the following applications:
- PURExpress,
- Toxic Protein Expression,
- Cell-Free Protein Expression,
- High-throughput cloning and automation solutions,
- Expression of Difficult Proteins,
- Disulfide-bonded Protein Expression, Protein Expression
Kit Components
Kit Components
The following reagents are supplied with this product:
| NEB # | Component Name | Component # | Stored at (°C) | Amount | Concentration | |
|---|---|---|---|---|---|---|
Properties & Usage
Application Features
- Quickly generate analytical amounts of protein for further characterization
- Confirmation of open reading frames
- Examination of the effects of mutations on ORFs
- Generation of truncated proteins to identify active domains and functional residues
- Introduction of modified, unnatural or labeled amino acids
- Epitope mapping
- Expression of toxic proteins
- Ribosome display
- Translation and/or protein folding studies
- In vitro compartmentalization
Related Products
Companion Products
Product Notes
- The DHFR control template is now supplied at 125 ng/µl. Use 2 µl for the positive control reaction. Template DNA, particularly plasmid DNA prepared by mini-prep (e.g. Qiagen) is often the major source of RNase contamination. We strongly recommend adding 20 units Murine RNase Inhibitor (NEB #M0314) to each reaction.
- PURExpress DHFR Control Template sequence files: Fasta GenBank
- Storage: All kit components should be stored at -80°C.
- Add Solution B to Solution A, do not dilute Solution B unbuffered. We recommend a starting amount of 250 ng template DNA per 25 μl reaction. The optimal amount of input DNA can be determined by setting up multiple reactions and titrating the amount of template DNA added to the reaction. Typically, the optimal amount will fall in a range of 25–1000 ng template per 25 μl reaction.
References
- Asahara, H. and Chong, S. (2010). In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of RNA polymerase holoenzyme. Nuc.Acid. Res.
- Desai, Bijoy J., Yuki Goto, Alessandro Cembran, Alexander A. Fedorov, Steven C. Almo, Jiali Gao, Hiroaki Suga, and John A. Gerlt (2014). Investigating the role of a backbone to substrate hydrogen bond in OMP decarboxylase using a site-specific amide to ester substitution. Proceedings of the National Academy of Sciences. 201411772.
- Gu, Liangcai, Chao Li, John Aach, David E. Hill, Marc Vidal, and George M. Church (2014). Multiplex single-molecule interaction profiling of DNA-barcoded proteins. Nature.
- Gupta, Pulkit, Shanmugapriya Sothiselvam, Nora Vázquez-Laslop, and Alexander S. Mankin (2013). Deregulation of translation due to post-transcriptional modification of rRNA explains why erm genes are inducible. Nature communications. 4,
- Kaiser, Christian M., Daniel H. Goldman, John D. Chodera, Ignacio Tinoco, and Carlos Bustamante (2011). The ribosome modulates nascent protein folding. Science. 334 (6063), 1723-1727.
- Nakagawa, So, Stephen S. Gisselbrecht, Julia M. Rogers, Daniel L. Hartl, and Martha L. Bulyk (2013). DNA-binding specificity changes in the evolution of forkhead transcription factors. Proceedings of the National Academy of Sciences. 110(30), 12349-12354.
- Ramadoss, Nitya S., John N. Alumasa, Lin Cheng, Yu Wang, Sharon Li, Benjamin S. Chambers, Hoon Chang et al (2013). Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proceedings of the National Academy of Sciences. 110(25), 10282-10287.
- Rosenblum, Gabriel, and Barry S. Cooperman (2014). Engine out of the chassis: Cell-free protein synthesis and its uses. FEBS letters. 588(2), 261-268.
- Stafford, Ryan L., Marissa L. Matsumoto, Gang Yin, Qi Cai, Juan Jose Fung, Heather Stephenson, Avinash Gill et al (2014). In vitro Fab display: a cell-free system for IgG discovery. Protein Engineering Design and Selection. 27(4), 97-109.
- Tuckey, Corinna, Haruichi Asahara, Ying Zhou, and Shaorong Chong (2014). Protein Synthesis Using a Reconstituted Cell‐Free System. Current Protocols in Molecular Biology. 16-31.
- Weirauch, Matthew T., Atina Cote, Raquel Norel, Matti Annala, Yue Zhao, Todd R. Riley, Julio Saez-Rodriguez et al (2013). Evaluation of methods for modeling transcription factor sequence specificity. Nature biotechnology. 31(2), 126-134.
- Noto, T., Kurth, H., Kataoka, K., Aronica, L., DeSouza, L., Siu, K., Pearlman, R., Gorovsky, M. and Mochizuki, K. (2010). The tetrahymena argonaute-binding protein Giw1p directs a mature argonaute-siRNAcomplex to the nucleus. Cell. 140, 692-703.
- Tanner, D., Cariello, D., Woolstenhulme, C., Broadbent, M. and Buskirk, A. (2009). Genetic identification of nascent peptides that induce ribosome stalling. J. Biol.Chem. 284, 34809-34818.
- Talabot-Ayer, D., Lamacchia, C., Gabay, C., and Palmer, G. (2009). Interleukin-33 is biologically activeindependently of Caspase-1 cleavage. J. Biol.Chem. 284, 19420-19426.
- Feng, Y. and Cronan, J. E. (2009). A new memberof the Eschericia coli fad regulon: transcriptional regulation offadM (ybaW). J.Bacteriol. 191, 6320-6328.
- Solaroli, N., Panayiotou, C., Johansson, M., and Karlsson, A. (2009). Identification of two active functional domains of human adenylate kinase 5. FEBSLett. 283, 2872-2876.
- Arenz, Stefan, Haripriya Ramu, Pulkit Gupta, Otto Berninghausen, Roland Beckmann, Nora Vázquez-Laslop, Alexander S. Mankin, and Daniel N. Wilson (2014). Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nature communications. 5,
- Chong, Shaorong (2014). Overview of Cell‐Free Protein Synthesis: Historic Landmarks, Commercial Systems, and Expanding Applications. Current Protocols in Molecular Biology. 16-30.
- Daugherty, Ashley B., Sridhar Govindarajan, and Stefan Lutz (2013). Improved Biocatalysts from a Synthetic Circular Permutation Library of the Flavin-Dependent Oxidoreductase Old Yellow Enzyme. Journal of the American Chemical Society . 334 (38), 14425-14432.
Protocols, Manuals & Usage
Protocols
- Protein Synthesis Reaction using PURExpress (E6800)
- Analysis of Synthesized Protein using PURExpress (E6800)
- Determination of Protein Synthesis Yield with PURExpress (E3313, E6800, E6840, E6850)
- Purification of Synthesized Protein using Reverse His-tag Purification
- Measurement of 35S-Methionine Incorporation by TCA Precipitation and Yield Determination using PURExpress
Manuals
The Product Manual includes details for how to use the product, as well as details of its formulation and quality controls.
Application Notes
Tools & Resources
Selection Charts
FAQs & Troubleshooting
FAQs
- When using PURExpress, I was unable to synthesize the control protein?
- When using PURExpress, I was able to synthesize the control protein, but the target sample is not present or present in low yield?
- When using PURExpress, I was able to synthesize the target protein, but full-length product is not major species?
- Where can I find many more detailed FAQs for PURExpress?
- Are there PURExpress citations?
Tech Tips
Thoroughly mix solutions A and B before using. Do not vortex Solution B or ribosomes, mix gently.
Solution A may have a cloudy white appearance. Add to the reaction as a uniform suspension.
Assemble the reactions in the following order on ice: Solution A, Solution B, RNAse Inhibitor, Water, Template DNA or RNA
Once reaction is assembled take time to make sure everything is thoroughly mixed by gently pipetting up and down, pulse spin and place at 37C for 2 to 4 hours.
Citations & Technical Literature
Citations
Additional Citations
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- E6800L_v1_10017429
- E6800S_v1_10017424
- E6800L_v1_10019041
- E6800L_v1_10021789
- E6800S_v1_10021788
- E6800L_v1_10022940
- E6800S_v1_10022722
- E6800L_v1_10029440
- E6800S_v1_10029441
- E6800L_v1_10032610
- E6800S_v1_10032609
- E6800L_v1_10036625
- E6800L_v1_10036913
- E6800S_v1_10036914
- E6800L_v1_10041043
- E6800L_v1_10043101
- E6800S_v1_10043102
- E6800S_v1_10043103
- E6800S_v1_10045359
- E6800L_v1_10045360
- E6800S_v1_10050086
- E6800L_v1_10051956
- E6800S_v1_10051957
- E6800S_v1_10051942
- E6800L_v1_10058459
- E6800S_v1_10058441
- E6800S_v1_10058460
- E6800L_v1_10063370
- E6800L_v1_10063391
- E6800L_v1_10069173
- E6800L_v1_10069959
- E6800S_v1_10069342
- E6800L_v1_10070109
- E6800S_v1_10069960
- E6800L_v1_10070932
- E6800L_v1_10073040
- E6800S_v1_10070931
- E6800L_v1_10077425
- E6800S_v1_10073038
- E6800L_v1_10080106
- E6800S_v1_10080107
- E6800L_v1_10083961
- E6800L_v1_10085455
- E6800L_v1_10086969
- E6800S_v1_10085456
- E6800L_v1_10087118
- E6800L_v1_10091924
- E6800S_v1_10097208
- E6800L_v2_10099566
- E6800S_v2_10100875
- E6800L_v2_10103789
- E6800L_v2_10107132
- E6800S_v2_10103790
- E6800L_v2_10110028
- E6800S_v2_10110029
- E6800S_v2_10119938
- E6800L_v2_10119940
- E6800S_v2_10122160
- E6800L_v2_10122167
- E6800L_v2_10127368
- E6800L_v2_10131882
- E6800S_v2_10131880
- E6800L_v2_10139804
- E6800L_v2_10141621
- E6800S_v2_10141620
- E6800L_v2_10144757
- E6800S_v2_10144753
- E6800S_v2_10144784
- E6800L_v2_10149825
- E6800S_v2_10151067
- E6800L_v2_10154181
- E6800L_v2_10156375
- E6800S_v2_10156374
- E6800L_v2_10158929
- E6800S_v2_10158927
- E6800L_v2_10171683
- E6800S_v2_10171682
- E6800S_v2_10174077
- E6800L_v2_10174076
- E6800L_v2_10176105
- E6800L_v2_10183088
- E6800S_v2_10183089
- E6800S_v2_10185167
- E6800L_v2_10185165
- E6800L_v2_10187115
- E6800S_v2_10187116
- E6800L_v2_10194580
- E6800S_v2_10191411
- E6800L_v2_10195964
- E6800S_v2_10198163
- E6800L_v2_10198162
- E6800S_v2_10206173
- E6800L_v2_10206174
- E6800S_v2_10209946
- E6800S_L_v1_0221805
- E6800L_v2_10220868
- E6800S_v2_10220861
- E6800L_v2_10222369
- E6800S_v2_10225999
- E6800S_v2_10230686
- E6800L_v2_10226197
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.PURExpress Solution A
Solution B (75 μl)
Control (DHFR) template (10 μl)
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email [email protected].This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Licenses
PURExpress® is based on the PURE System Technology originally developed by Dr. Takuya Ueda at the University of Tokyo and commercialized as the PURESYSTEM® by BioComber (Tokyo, Japan).Licensed from BioComber (Tokyo, Japan) under Patent Nos. 7,118,883; WO2005-105994 and JP2006-340694. For research use only. Commercial use of PURExpress® In vitro Protein Synthesis Kit requires a license from New England Biolabs, Inc. This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.