NEBExpress® Competent E. coli (High Efficiency)
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
C2523I |
NEB Express Competent E. coli (High Efficiency) |
6 x 0.2 ml | - | Unavailable in your region | |
C2523H |
NEB Express Competent E. coli (High Efficiency) |
20 x 0,05 ml | - | Unavailable in your region |
NEBExpress® Competent E. coli (High Efficiency)
Having trouble opening your tubes? NEB Tube Opener can help!
Product Introduction
High efficiency chemically competent E. coli cells suitable for non-T7 protein expression.
- Deficient in proteases Lon and OmpT
- Resistant to phage T1 (fhuA2)
- Free of animal products
- No dry ice surcharge on competent cell shipments
| Catalog # | Size | Concentration |
|---|---|---|
| C2523H | 20 x 0.05 ml | |
| C2523I | 6 x 0.2 ml |
- Product Information
- Protocols, Manuals & Usage
- Tools & Resources
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
Highlights
- Enhanced BL21 derivative ideal for Plac, Ptac, Ptrc expression vectors
- Deficient in proteases Lon and OmpT
- Resistant to phage T1 (fhuA2)
- Does not restrict methylated DNA (McrA-, McrBC-, EcoBr-m-, Mrr-)
- Free of animal products
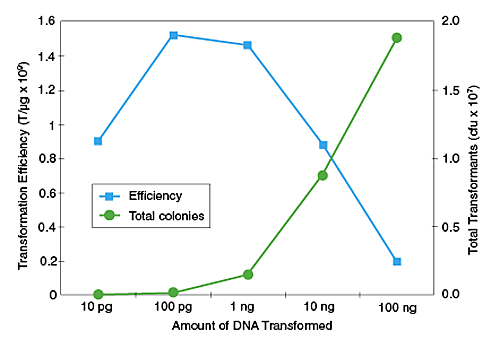
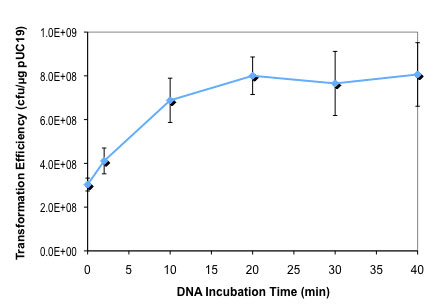
Transformation Efficiency
0.6-1 x 109 cfu/μg pUC19 DNAGenotype
fhuA2 [lon] ompT gal sulA11 R(mcr-73::miniTn10--TetS)2 [dcm] R(zgb-210::Tn10--TetS) endA1 Δ(mcrC-mrr)114::IS10- This product is related to the following categories:
- Non-T7 Expression,
- E. coli Protein Expression Strains,
- Protein Expression,
- E. coli Expression Strains,
- This product can be used in the following applications:
- Non-T7 Expression,
- Protein Expression in E. Coli,
- Protein Expression
Reagents Supplied
Reagents Supplied
The following reagents are supplied with this product:
| NEB # | Component Name | Component # | Stored at (°C) | Amount | Concentration | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
Properties & Usage
Antibiotic for Plasmid Selection
| Antibiotics for Plasmid Selection | Working Concentration |
|---|---|
| Ampicillin | 100 µg/ml |
| Carbenicillin | 100 µg/ml |
| Chloramphenicol | 33 µg/ml |
| Kanamycin | 30 µg/ml |
| Tetracycline | 15 µg/ml |
| Streptomycin | 25 µg/ml |
Shipping Notes
- Ships on dry ice
Antibiotic Resistance
- nit
Features
- Versatile non-T7 expression strain
- Protease deficient
Application Features
Chemically competent E. coli cells suitable for high efficiency transformation and protein expression. Recommended host strain for pMAL Protein Fusion and Purification System.

Related Products
Companion Products
Materials Sold Separately
Product Notes
- CAUTION: This product contains DMSO, a hazardous material. Review the MSDS before handling.
- STORAGE AND HANDLING: Competent cells should be stored at -80°C. Storage at -20°C will result in a significant decrease in transformation efficiency. Cells lose efficiency whenever they are warmed above -80°C, even if they do not thaw.
Protocols, Manuals & Usage
Protocols
Usage & Guidelines
Application Notes
Tools & Resources
Selection Charts
FAQs & Troubleshooting
FAQs
- Does New England Biolabs offer strains of Competent E.coli suited for protein expression?
- How long should I incubate cells on ice after DNA has been added (NEB #C2523H and NEB #C2523I)?
- What are the solutions/recipes (C2523)?
- What are the strain properties (C2523)?
- What is the difference between NEB #C2523H and NEB #C2523I?
- Why are there no colonies or no growth in liquid culture (C2523)?
- Why is induced protein insoluble (C2523)?
- Why is there no protein visible on gel or no activity (C2523)?
- Can I store competent cells at -20°C instead of -80°C?
- Can the NEBExpress Competent E.coli (High Efficiency) be used for the expression of constructs containing a T7 promoter?
- What is the optimal heat shock time for this strain (NEB #C2523H and NEB #C2523I)?
- Which kind of transformation tubes should be used?
- What volume of DNA can be added into competent cells?
- What is the shelf life for this strain (NEB #C2523H and NEB #C2323I)?
- Are NEB's competent cells compatible with the “Mix & Go" protocol?
- How should I store SOC Outgrowth Medium? The SOC I received with my competent cells recommends storage at either room temperature or 4°C, however, when I purchase it as a stand alone product, it recommends storing it at 4°C. Which is better?
Citations & Technical Literature
Citations
Additional Citations
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- C2523H_I_v1_0161606
- C2523H_I_v1_0151606
- C2523H_I_v1_0151608
- C2523H_I_v1_0151609
- C2523H_I_v1_0151611
- C2523H_I_v1_0161703
- C2523H_I_v1_0151704
- C2523H_I_v1_0161705
- C2523H_I_v1_0161708
- C2523H_I_v1_0161710
- C2523H_I_v1_0161801
- C2523H_I_v1_0161802
- C2523H_I_v1_0161803
- C2523I_v1_10012782
- C2523H_v1_10016257
- C2523I_v1_10017217
- C2523H_v1_10020863
- C2523H_v1_10026187
- C2523H_v1_10028613
- C2523I_v1_10032356
- C2523H_v1_10032819
- C2523H_v1_10036765
- C2523H_v1_10038798
- C2523I_v1_10039039
- C2523I_v1_10042694
- C2523H_v1_10046062
- C2523I_v1_10045593
- C2523I_v1_10048230
- C2523H_v1_10049343
- C2523I_v1_10050605
- C2523H_v1_10051588
- C2523I_v1_10052486
- C2523H_v1_10056796
- C2523H_v1_10058653
- C2523I_v1_10059032
- C2523H_v1_10062452
- C2523I_v1_10064426
- C2523I_v1_10066977
- C2523I_v1_10068104
- C2523I_v1_10069275
- C2523H_v2_10072127
- C2523I_v2_10072423
- C2523I_v2_10077689
- C2523H_v2_10079072
- C2523H_v2_10083739
- C2523I_v2_10082103
- C2523I_v2_10086603
- C2523H_v2_10087091
- C2523I_v2_10090670
- C2523I_v2_10096413
- C2523H_v2_10098360
- C2523I_v2_10100224
- C2523H_v2_10102092
- C2523I_v2_10102720
- C2523I_v2_10105519
- C2523H_v2_10108144
- C2523I_v2_10108145
- C2523I_v2_10113186
- C2523H_v2_10116425
- C2523I_v2_10115833
- C2523I_v2_10119757
- C2523I_v2_10122542
- C2523I_v2_10123639
- C2523H_v2_10122541
- C2523I_v2_10128276
- C2523H_v2_10129888
- C2523I_v2_10135794
- C2523H_v2_10137579
- C2523I_v2_10139601
- C2523I_v2_10142458
- C2523I_v2_10145531
- C2523H_v2_10147009
- C2523I_v2_10149437
- C2523H_v2_10151667
- C2523I_v2_10152257
- C2523I_v2_10159675
- C2523H_v2_10160823
- C2523I_v2_10162449
- C2523H_v2_10163242
- C2523I_v2_10166315
- C2523H_v2_10169178
- C2523H_v2_10173582
- C2523I_v2_10171989
- C2523H_v2_10176534
- C2523H_v2_10178252
- C2523I_v2_10182036
- C2523I_v2_10188906
- C2523H_v2_10187588
- C2523H_v2_10191328
- C2523H_v2_10195829
- C2523I_v2_10195274
- C2523H_v2_10204044
- C2523I_v2_10206194
- C2523H_v2_10208370
- C2523I_v2_10210989
- C2523I_v2_10212396
- C2523H_v2_10213907
- C2523I_v2_10225944
- C2523H_v2_10220884
- C2523I_v2_10231070
- C2523H_v2_10232238
- C2523I_v2_10236690
- C2523H_v2_10239114
- C2523I_v2_10240005
- C2523H_v2_10244999
- C2523I_v2_10245648
- C2523H_v2_10248370
- C2523H_v2_10251035
- C2523I_v2_10250925
- C2523H_v2_10256112
- C2523H_v2_10257189
- C2523I_v2_10261001
- C2523H_v2_10267678
- C2523I_v2_10267200
- C2523H_v2_10269518
- C2523H_v2_10273614
- C2523H_v2_10274156
- C2523I_v2_10274532
- C2523I_v2_10280510
- C2523H_v2_10280914
- C2523I_v2_10288603
- C2523H_v2_10292315
- C2523H_v2_10301997
- C2523I_v2_10302492
- C2523H_v2_10309810
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.NEBExpress® Competent E. coli (High Efficiency)
SOC Outgrowth Medium
pUC19 Vector
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email [email protected].This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.
Protocol
Expression Using NEB Express (C2523)
Protocol
5 Minute Transformation (C2523)
Protocol
5 Minute Transformation (C2523)
Protocol
5 Minute Transformation (C2523)
Protocol
5 Minute Transformation (C2523)
Protocol
Expression Using NEB Express (C2523)
Protocol
Expression Using NEB Express (C2523)
Protocol
Expression Using NEB Express (C2523)


