HiScribe® T7 High Yield RNA Synthesis Kit
Product information| Code | Name | Size | Quantity | Price | |
|---|---|---|---|---|---|
E2040S |
T7 High Yield RNA Synthesis Kit |
50 rxns | - | Unavailable in your region |
HiScribe® T7 High Yield RNA Synthesis Kit
Now includes separate tube of DTT
Product Introduction
- Up to 180 μg of RNA per reaction from 1 μg of control template
- Enables full substitution of NTPs for labeling and incorporation of modified bases
- Linearized control template included for verification of RNA synthesis
- Getting ready to scale up RNA synthesis? Download our new technical note “Scaling of High-Yield In vitro Transcription Reactions for Linear Increase of RNA Production” for a generalized set of recommendations for synthesizing high yields of RNA.
| Catalog # | Size | Concentration |
|---|---|---|
| E2040S | 50.0 reactions |
Featured Videos
View Video Library- Product Information
- Protocols, Manuals & Usage
- FAQs & Troubleshooting
- Citations & Technical Literature
- Quality, Safety & Legal
- Other Products You May Be Interested In
Product Information
Description
The HiScribe T7 High Yield RNA Synthesis Kit is an extremely flexible system for in vitro transcription of RNA using T7 RNA Polymerase. The kit allows for synthesis many kinds of RNA including internally labeled and co-transcriptionally capped transcripts.RNA synthesized from the kit is suitable for many applications including RNA structure and function studies, ribozyme biochemistry, probes for RNase protection assays and hybridization based blots, anti-sense RNA and RNAi experiments, microarray analysis, microinjection, and in vitro translation and RNA vaccines.
The kit contains sufficient reagents for 50 reactions of 20 μl each. Each standard reaction yields up to 180 μg of RNA from 1 μg control template. Each kit can yield up to 9 mg RNA. For 32P labeling, the kit contains enough reagents for 100 reactions of 20 μl each.
Materials Not Included:
- DNA Template: The DNA template must be linear and contain the T7 RNA Polymerase promoter with correct orientation in relation to target sequence to be transcribed.
- 3′-O-Me-m7G(5′)ppp(5′)G RNA Cap Structure Analog (NEB #S1411)
- m7G(5′)ppp(5′)G RNA Cap Structure Analog (NEB #S1404)
- m7G(5′)ppp(5′)A RNA Cap Structure Analog (NEB #S1405)
- G(5′)ppp(5′)A RNA Cap Structure Analog (NEB #S1406)
- G(5′)ppp(5′)G RNA Cap Structure Analog (NEB #S1407)
- Modified-NTP: Biotin-, Fluorescein-, Digoxigenin-, or Aminoallyl-NTP
- Labeling: [α-32P] labeled ribonucleotide (800-6,000 Ci/mmol)
- General: 37°C incubator or PCR machine, nuclease-free water
- DNase I: DNase I (RNase-free) (NEB #M0303)
- Purification: Buffer- or water-saturated phenol/chloroform, ethanol and 3 M sodium acetate, pH 5.2, spin columns
- Gel Analysis: Gels and running buffers, gel apparatus, power supply
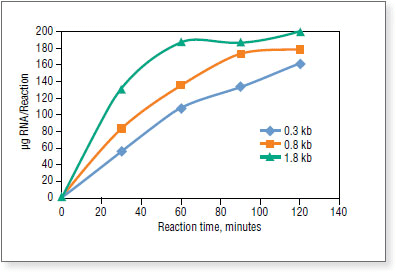
Reactions were incubated at 37°C in a PCR machine. Transcripts were purified by spin columns and quantified on NanoDrop™ Spectrophotometer.
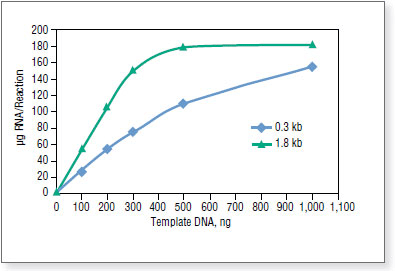
Standard reactions were incubated at 37°Cin a PCR machine for 2 hours. Transcripts were purified by spin columns and quantified on NanoDrop™ Spectrophotometer.
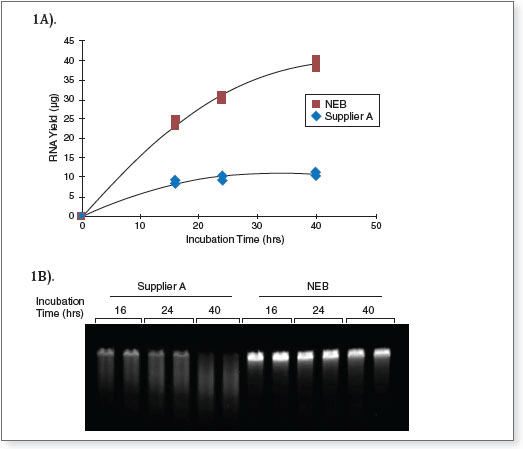
reactions were assembled, in duplicate, according to the manufacturers’ suggested protocols using 3 ng of dsDNA template encoding a 1.8 kb RNA, and incubated at 37°C for 16, 24 and 40 hours. At each time point, the corresponding tubes were transferred to -20°C to stop the reaction. Transcription reactions were column purified after the last time point.
(A) Transcript yield – After column purification, RNA concentration was measured using a NanoDrop spectrophotometer and total RNA yield was calculated. These data demonstrate that a substantially higher yield of RNA was synthesized using the HiScribe T7 High Yield RNA Synthesis Kit as compared to the competitor’s kit.
(B) Transcript integrity – 150 ng of column purified RNA was run a 1.2% denaturing agarose gel, stained with ethidium bromide and visualized by UV fluorescence. The data demonstrate greatly improved transcript integrity after extended duration RNA synthesis reactions using the HiScribe T7 High Yield RNA Synthesis Kit as compared to the competitor’s kit.
- This product is related to the following categories:
- RNA Capping,
- RNA Synthesis In vitro Transcription (IVT),
Kit Components
Kit Components
The following reagents are supplied with this product:
Properties & Usage
Related Products
Companion Products
- RNA Loading Dye, (2X)
- RNase Inhibitor, Human Placenta
- RNase Inhibitor, Murine
- DNase I (RNase-free)
- Q5® Hot Start High-Fidelity DNA Polymerase
- ssRNA Ladder
- Low Range ssRNA Ladder
- 3´-0-Me-m7G(5')ppp(5')G RNA Cap Structure Analog
- m7G(5')ppp(5')A RNA Cap Structure Analog
- G(5')ppp(5')A RNA Cap Structure Analog
- G(5')ppp(5')G RNA Cap Structure Analog
- m7G(5')ppp(5')G RNA Cap Structure Analog
- Vaccinia Capping System
- mRNA Cap 2 O Methyltransferase
- E. coli Poly(A) Polymerase
- Ribonucleotide Solution Mix
- Ribonucleotide Solution Set
Protocols, Manuals & Usage
Protocols
- DNA Template Preparation (E2040)
- RNA Synthesis with Modified Nucleotides (E2040)
- Purification of Synthesized RNA (E2040)
- Standard RNA Synthesis (E2040)
- Capped RNA Synthesis (E2040)
- High Specific Activity Radiolabeled RNA Probe Synthesis (E2040)
- Evaluation of Reaction Products (E2040)
- Poly(A) Tailing of RNA using E. coli Poly(A) Polymerase (NEB# M0276)
- Protocol for Co-transcriptional capping using CleanCap® Reagent AG from TriLink and HiScribe™ T7 High Yield RNA Synthesis Kit from New England Biolabs®
Manuals
Application Notes
FAQs & Troubleshooting
FAQs
Troubleshooting
Control Reaction
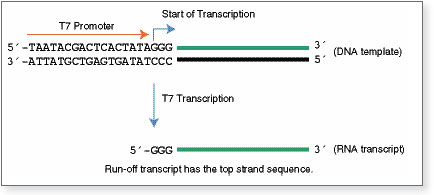
The FLuc control template DNA is a linearized plasmid containing the firefly luciferase gene under the transcriptional control of T7 promoter. The size of the runoff transcript is 1.8 kb. The control reaction should yield ≥ 150 μg RNA transcript in 2 hours.
If the control reaction is not working, there may be technical problems during reaction set up. Repeat the reaction by following the protocol carefully; take any precaution to avoid RNase contamination. Contact NEB for technical assistance.
The control plasmid sequence can be found within the DNA Sequences and Maps Tool under the name "FLuc Control Plasmid". The FLuc control template is generated by linearizing the plasmid with StuI.
Low Yield of Full-length RNA
If the transcription reaction with your template generates full-length RNA, but the yield is significantly lower than expected, it is possible that contaminants in the DNA template are inhibiting the RNA polymerase, or the DNA concentration may be incorrect. Alternatively, additional purification of DNA template may be required. Phenol-chloroform extraction is recommended (see template DNA preparation section).
Low Yield of Short Transcript
High yields of short transcripts (< 0.3 kb) are achieved by extending incubation time and increasing the amount of template. Incubation of reactions up to 16 hours (overnight) or using up to 2 μg of template will help to achieve maximum yield.
RNA Transcript Smearing on Denaturing Gel
If the RNA appears degraded (e.g. smeared) on denaturing agarose or polyacrylamide gel, DNA template is contaminated with RNase. DNA templates contaminated with RNase can affect the length and yield of RNA synthesized (a smear below the expected transcript length). If the plasmid DNA template is contaminated with RNase, perform phenol/chloroform extraction, then ethanol precipitate and dissolve the DNA in nuclease-free water (see template DNA preparation section).
RNA Transcript of Larger Size than Expected
If the RNA transcript appears larger than expected on a denaturing gel, template plasmid DNA may be incompletely digested. Even small amounts of undigested circular DNA can produce large amounts of long transcripts. Check template for complete digestion, if undigested plasmid is confirmed, repeat restriction enzyme digestion.
Larger size bands may also be observed when the RNA transcript is not completely denatured due to the presence of strong secondary structures.
RNA Transcript of Smaller Size than Expected
If denaturing gel analysis shows the presence of smaller bands than the expected size, it is most likely due to premature termination by the polymerase. Some sequences which resemble T7 RNA Polymerase termination signals will cause premature termination. Incubating the transcription reaction at lower temperatures, for example at 30°C, may increase the proportion of full-length transcript, however the yield will be decreased. For GC rich templates, or templates with secondary structures, incubation at 42°C may improve yield of full-length transcript.
If premature termination of transcription is found in high specific activity radiolabeled RNA probe synthesis, increase the concentration of “limiting NTP”. Additional "cold" NTP can be added to the reaction to increase the proportion of full-length transcript, however the improvement in yield of full-length product will compromise the specific activity of the probe.
Tech Tips
It is important to mix each component well before setting up reactions.
Make sure reactions are thoroughly mixed.
We recommend incubating the reactions in a dry air incubator or in a PCR machine.
Citations & Technical Literature
Citations
Additional Citations
- Lee, NC., Larionov, V., Kouprina, N. (2015) Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast Nucleic Acids Res; 43(8), e55. PubMedID: 25690893
- Jaitin, DA., Kenigsberg, E., Keren-Shaul, H., Elefant, N., Paul, F., Zaretsky, I., Mildner, A., Cohen, N., Jung, S., Tanay, A. and Amit, I. (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science; 343, 776-779. PubMedID: 24531970
Quality, Safety & Legal
Quality Assurance Statement
Quality Control tests are performed on each new lot of NEB product to meet the specifications designated for it. Specifications and individual lot data from the tests that are performed for this particular product can be found and downloaded on the Product Specification Sheet, Certificate of Analysis, data card or product manual. Further information regarding NEB product quality can be found here.Specifications
The Specification sheet is a document that includes the storage temperature, shelf life and the specifications designated for the product. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]Certificate Of Analysis
The Certificate of Analysis (COA) is a signed document that includes the storage temperature, expiration date and quality controls for an individual lot. The following file naming structure is used to name these document files: [Product Number]_[Size]_[Version]_[Lot Number]- E2040S_v1_0241803
- E2040S_v1_10012595
- E2040S_v1_10013491
- E2040S_v1_10014774
- E2040S_v1_10018975
- E2040S_v1_10021740
- E2040S_v1_10022756
- E2040S_v1_10027773
- E2040S_v1_10031646
- E2040S_v1_10032241
- E2040S_v1_10035263
- E2040S_v1_10032035
- E2040S_v1_10041536
- E2040S_v1_10044351
- E2040S_v1_10045383
- E2040S_v1_10047695
- E2040S_v1_10050954
- E2040S_v1_10054479
- E2040S_v1_10056892
- E2040S_v1_10059199
- E2040S_v1_10061569
- E2040S_v1_10065045
- E2040S_v1_10068416
- E2040S_v1_10073088
- E2040S_v1_10078059
- E2040S_v1_10078861
- E2040S_v1_10082352
- E2040S_v1_10087061
- E2040S_v1_10090166
- E2040S_v1_10093284
- E2040S_v1_10098040
- E2040S_v1_10099587
- E2040S_v1_10100208
- E2040S_v1_10105808
- E2040S_v1_10109077
- E2040S_v1_10114318
- E2040S_v1_10116523
- E2040S_v1_10120183
- E2040S_v1_10123751
- E2040S_v1_10126799
- E2040S_v1_10130030
- E2040S_v1_10133564
- E2040S_v1_10135706
- E2040S_v1_10138225
- E2040S_v1_10139668
- E2040S_v1_10143402
- E2040S_v1_10146333
- E2040S_v1_10148493
- E2040S_v1_10153465
- E2040S_v1_10154863
- E2040S_v1_10155138
- E2040S_v1_10158087
- E2040S_v1_10161532
- E2040S_v1_10163354
- E2040S_v1_10165920
- E2040S_v1_10168659
- E2040S_v1_10169131
- E2040S_v1_10170335
- E2040S_v1_10172139
- E2040S_v1_10177737
- E2040S_v1_10178624
- E2040S_v1_10179975
- E2040S_v1_10183049
- E2040S_v1_10187108
- E2040S_v1_10187540
- E2040S_v1_10190217
- E2040S_v1_10193354
- E2040S_v1_10194679
- E2040S_v1_10195873
- E2040S_v1_10201919
- E2040S_v1_10202940
- E2040S_v2_10205871
- E2040S_v2_10207484
- E2040S_v2_10208121
- E2040S_v2_10211578
- E2040S_v2_10215041
- E2040S_v1_0251804
- E2040S_v2_10219195
- E2040S_v2_10222703
- E2040S_v2_10224853
- E2040S_v2_10227132
- E2040S_v3_10229427
- E2040S_v3_10232798
- E2040S_v3_10233999
- E2040S_v3_10234843
- E2040S_v3_10235841
- E2040S_v3_10238457
Safety DataSheets
The following is a list of Safety Data Sheet (SDS) that apply to this product to help you use it safely.CTP
FLuc Control Template
UTP
ATP
T7 RNA Polymerase Mix
GTP
10X T7 Reaction Buffer
Dithiothreitol (DTT)
Legal and Disclaimers
Products and content are covered by one or more patents, trademarks and/or copyrights owned or controlled by New England Biolabs, Inc (NEB). The use of trademark symbols does not necessarily indicate that the name is trademarked in the country where it is being read; it indicates where the content was originally developed. The use of this product may require the buyer to obtain additional third-party intellectual property rights for certain applications. For more information, please email [email protected].This product is intended for research purposes only. This product is not intended to be used for therapeutic or diagnostic purposes in humans or animals.
New England Biolabs (NEB) is committed to practicing ethical science – we believe it is our job as researchers to ask the important questions that when answered will help preserve our quality of life and the world that we live in. However, this research should always be done in safe and ethical manner. Learn more.
Other Products You May Be Interested In
The supporting documents available for this product can be downloaded below.